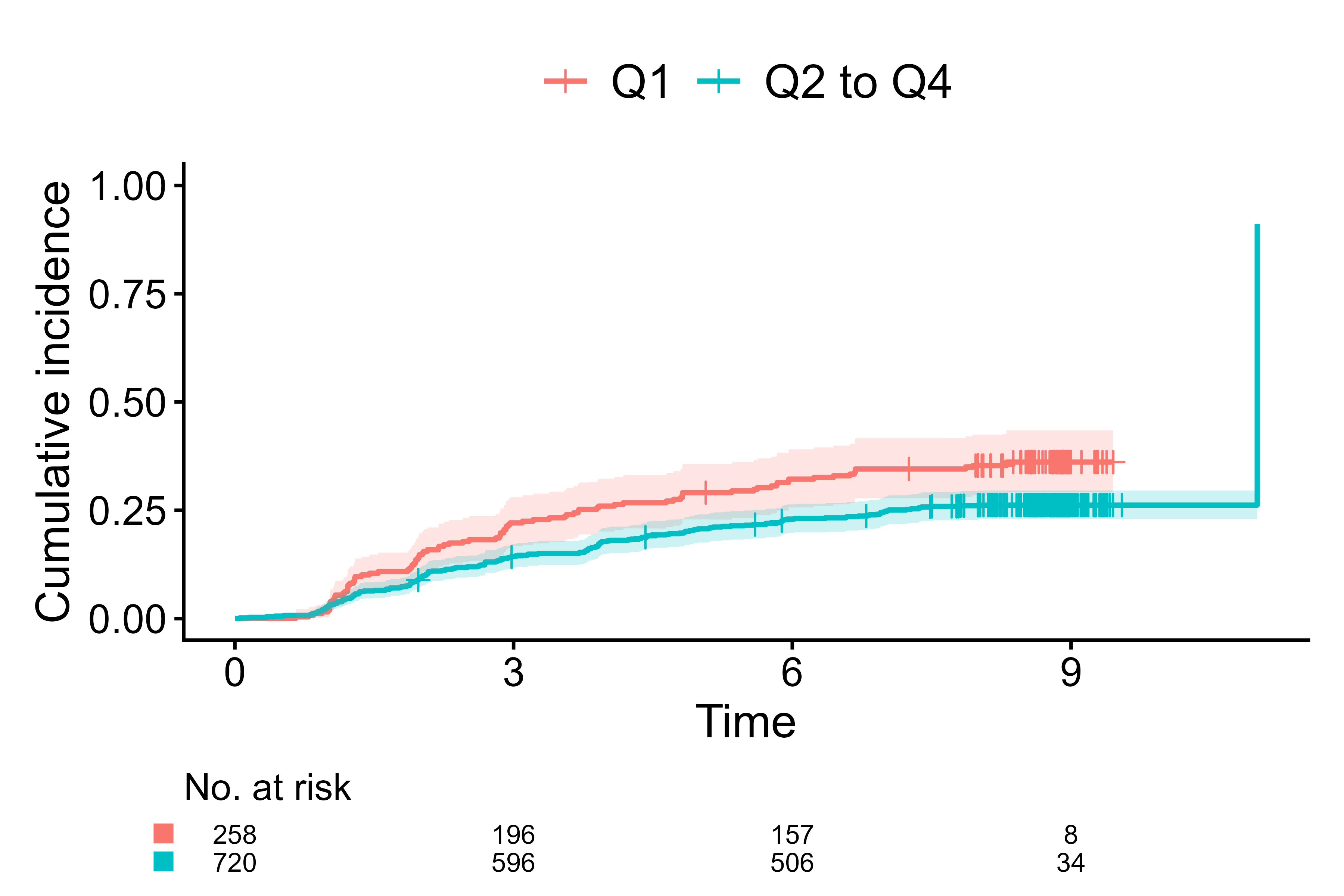
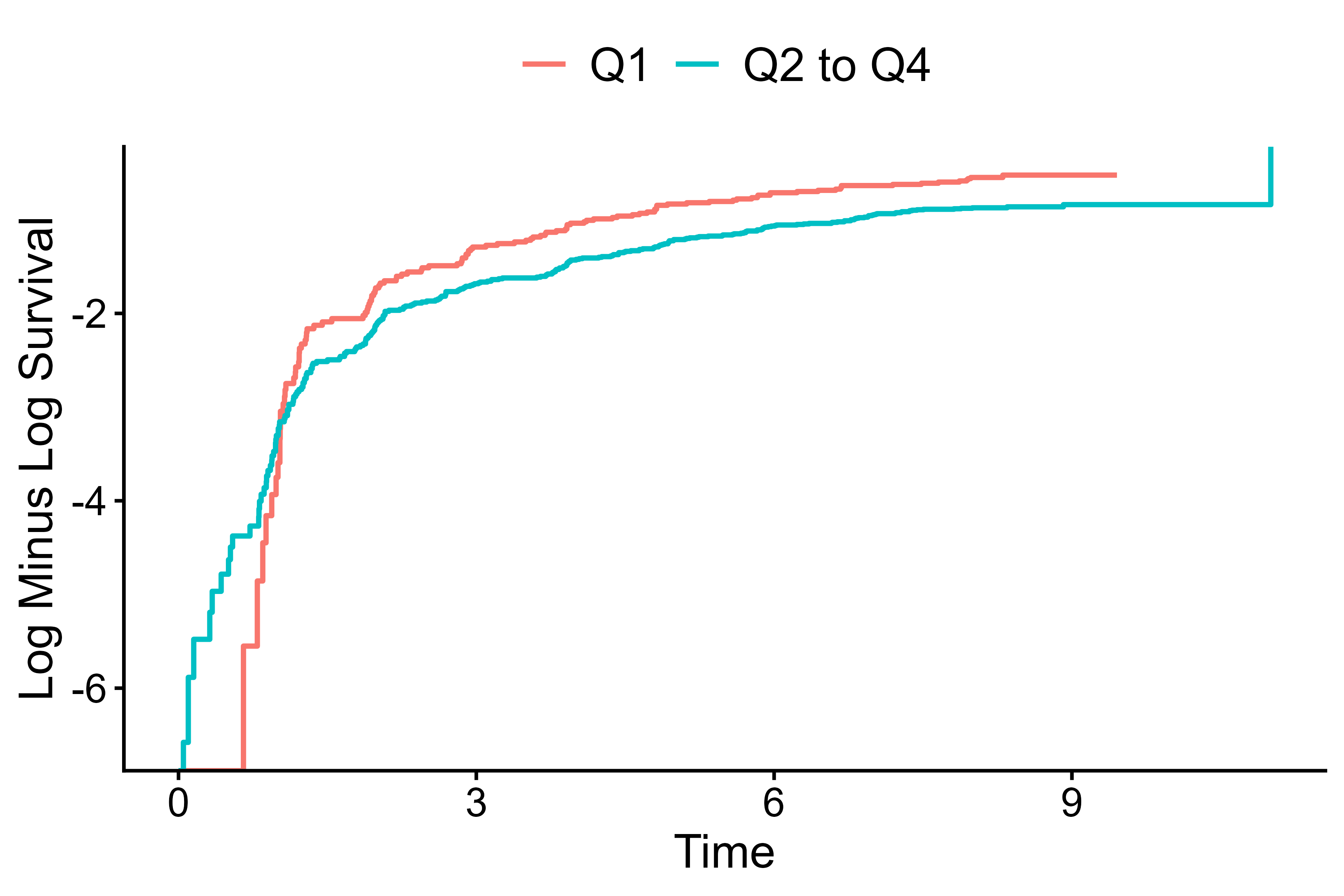
Default for survival data
data(diabetes.complications)
diabetes.complications$d <- as.integer(diabetes.complications$epsilon>0)
diabetes.complications$fruitq1 <- ifelse(
diabetes.complications$fruitq == "Q1","Q1","Q2 to Q4"
)
cifplot(Event(t,d) ~ fruitq1,
data = diabetes.complications,
outcome.type = "survival")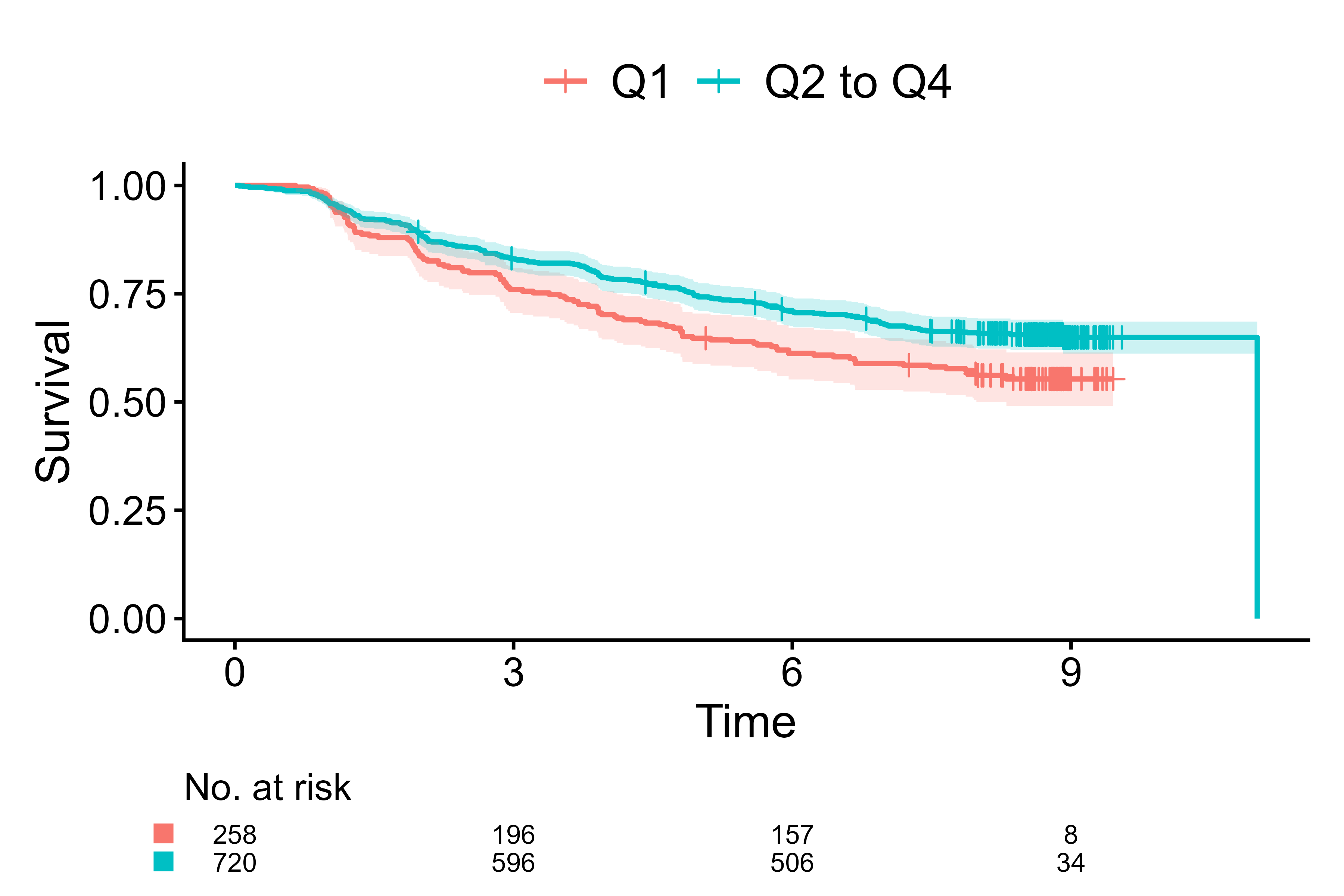
Default of cifplot() for survival data
Visual elements
cifplot(Event(t,epsilon) ~ fruitq1,
data = diabetes.complications,
outcome.type = "competing-risk",
add.conf = FALSE,
add.risktable = FALSE,
add.censor.mark = FALSE)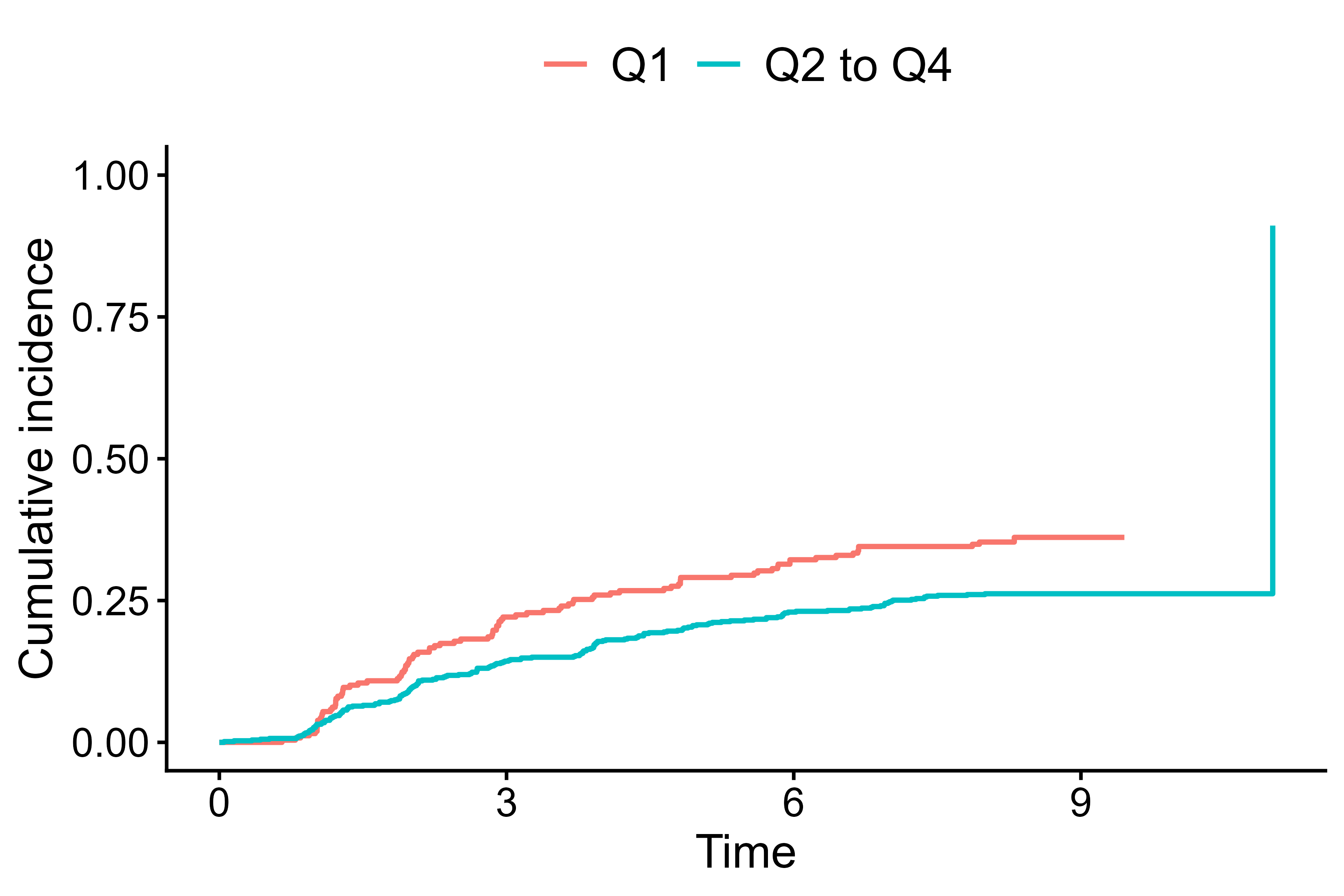
Hide confidence interval, risk table, and censor marks
cifplot(Event(t,epsilon) ~ fruitq1,
data = diabetes.complications,
outcome.type = "competing-risk",
add.risktable = FALSE,
add.estimate.table = TRUE,
add.censor.mark = FALSE,
add.competing.risk.mark = TRUE)
Add a table of CIF estimates and event-2 time marks
# Extract event-2 time from data frame
time_to_event <- extract_time_to_event(Event(t, epsilon) ~ fruitq1,
data = diabetes.complications,
which.event = "event2")
# Ensure named list by strata
stopifnot(is.list(time_to_event), length(time_to_event) > 0)
# Add event-2 time marks using external event-2 time
cifplot(Event(t, epsilon) ~ fruitq1,
data = diabetes.complications,
outcome.type = "competing-risk",
add.risktable = FALSE,
add.estimate.table = TRUE,
add.censor.mark = FALSE,
add.competing.risk.mark = TRUE,
competing.risk.time = time_to_event)
Add event-2 time marks using external event-2 time
cifplot(Event(t, epsilon) ~ fruitq1,
data = diabetes.complications,
outcome.type = "competing-risk",
add.risktable = FALSE,
add.estimate.table = TRUE,
symbol.risk.table = "circle",
add.censor.mark = FALSE,
add.competing.risk.mark = TRUE,
shape.competing.risk.mark = 1,
size.competing.risk.mark = 4,
competing.risk.time = time_to_event)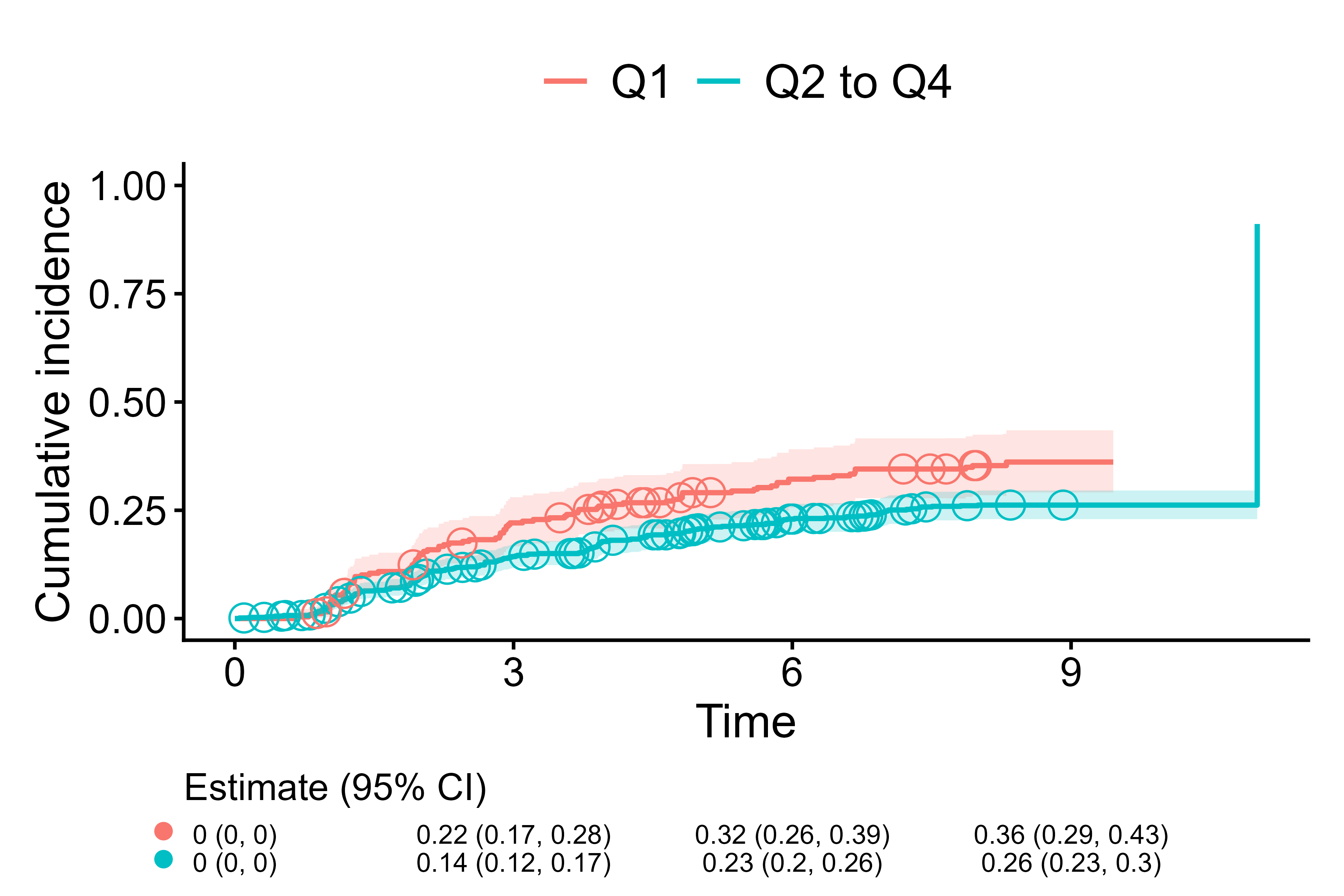
Use (open) circle for strata in estimate table and event-2 time marks
Risk ratio label
# Fit direct polytomous regression
output1 <- polyreg(nuisance.model=Event(t,epsilon)~1, exposure="fruitq1",
data=diabetes.complications, effect.measure1="RR", effect.measure2="RR",
time.point=8, outcome.type="competing-risk")
# Create a label of the risk ratio of event1
output2 <- effect_label.polyreg(output1,
event="event1",
add.exposure.levels=FALSE,
add.outcome=FALSE,
add.conf=FALSE)
output3 <- cifplot(Event(t,epsilon) ~ fruitq1,
data = diabetes.complications,
outcome.type = "competing-risk")
# Add the label
output4 <- output3$plot + ggplot2::annotate("text", x=6, y=0.8,
label=output2, hjust=1, vjust=1)
print(output4)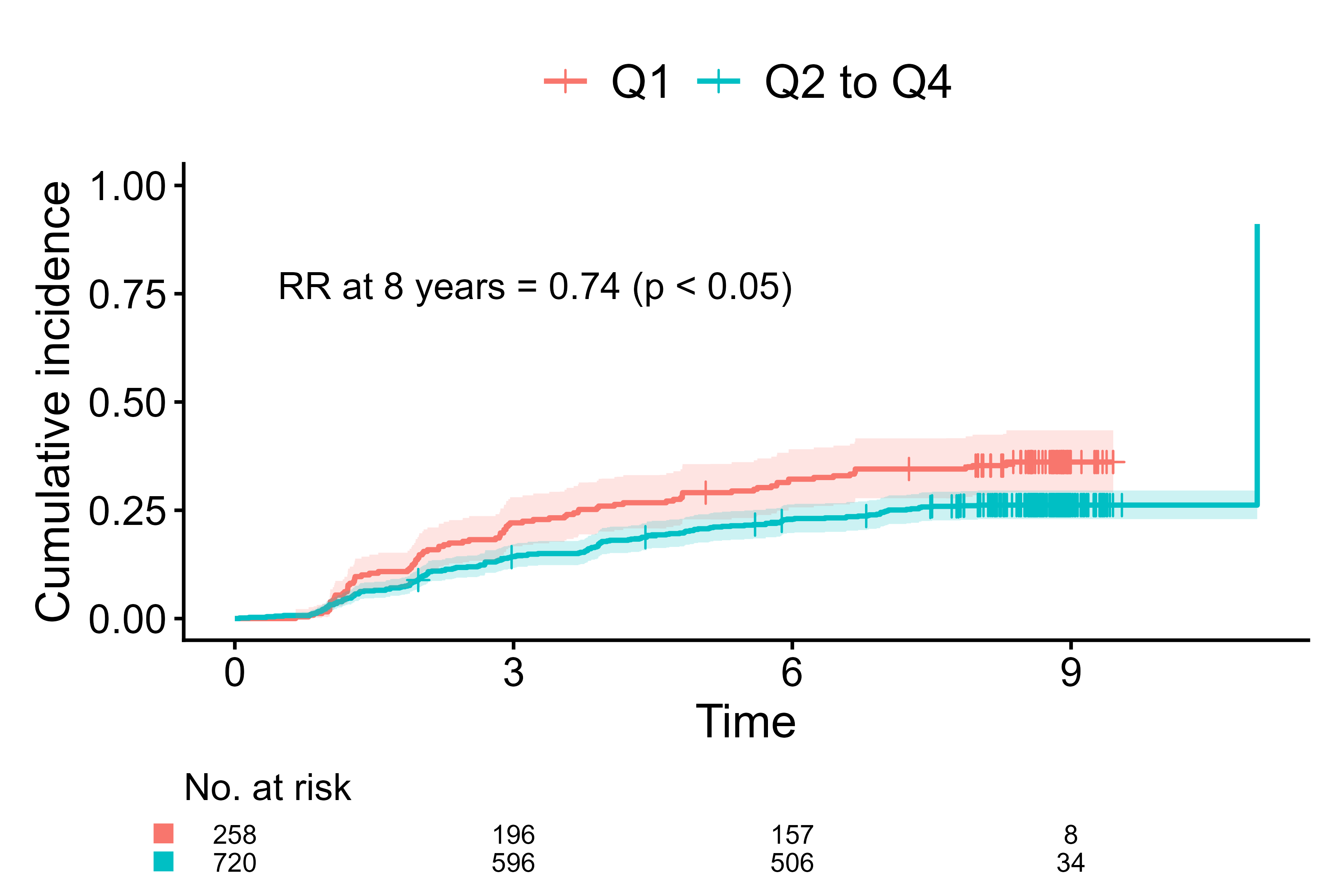
Add risk ratio label using polyreg() and effect_label.polyreg()
Panel mode
cifplot(Event(t,epsilon) ~ fruitq + fruitq1,
data = diabetes.complications,
outcome.type = "competing-risk",
panel.per.variable = TRUE)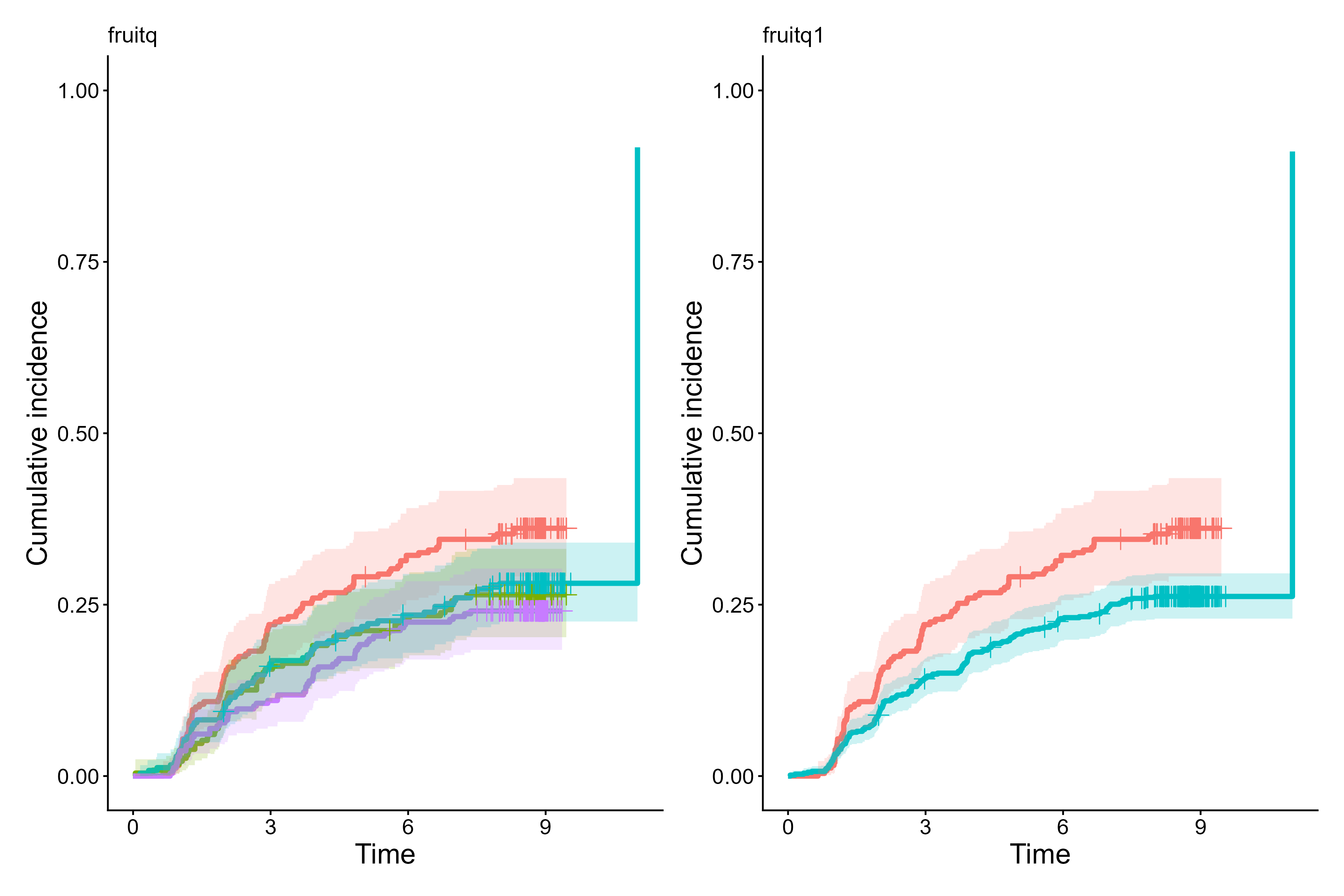
Cumulative incidence curves per each stratification variable
cifplot(Event(t,epsilon) ~ fruitq1,
data = diabetes.complications,
outcome.type = "competing-risk",
panel.per.event = TRUE)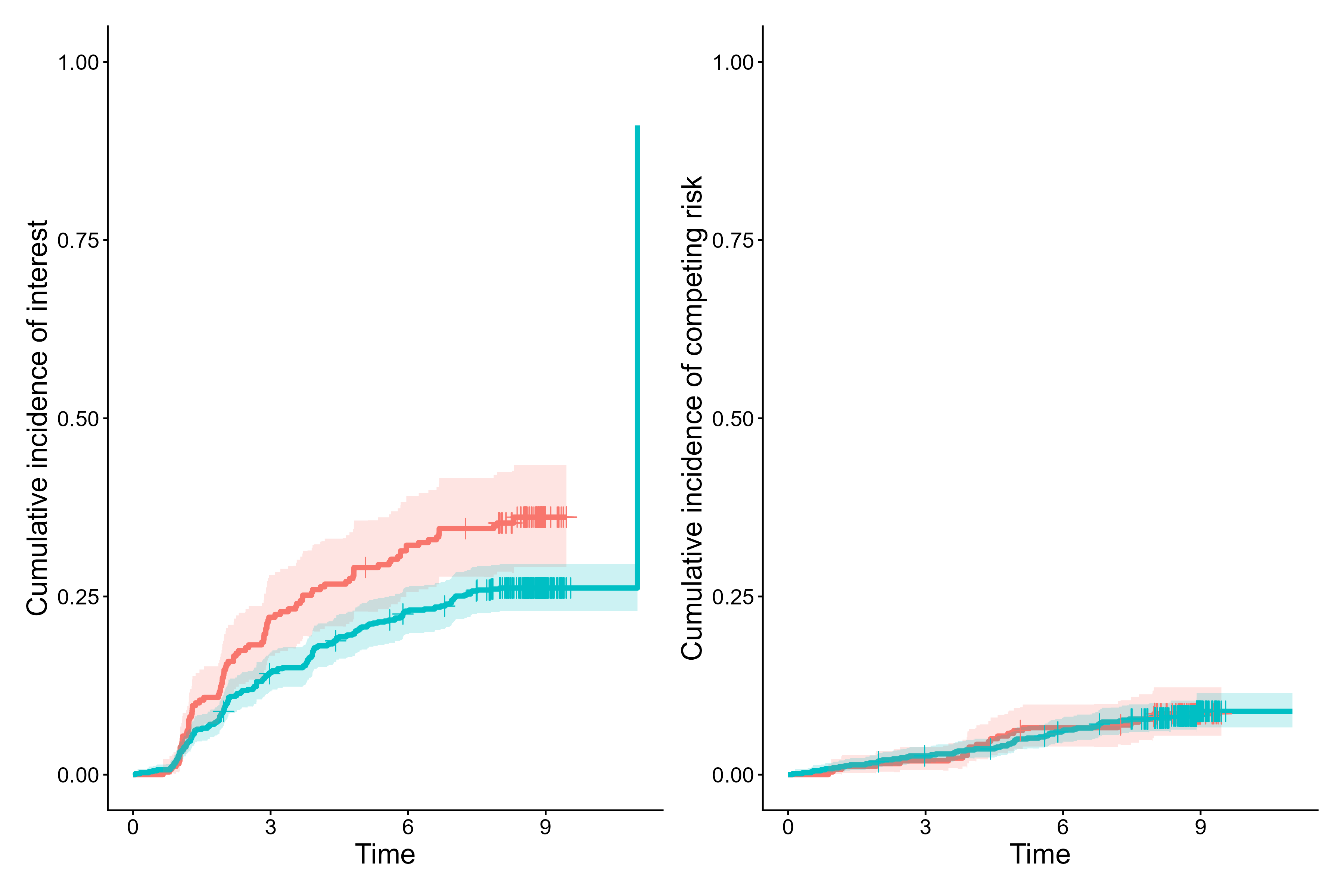
Cumulative incidence curves for event 1 vs event 2
cifplot(Event(t,d) ~ fruitq1,
data = diabetes.complications,
outcome.type = "survival",
panel.censoring = TRUE)
Survival curves for event vs censoring
Zoomed-in-view
output5 <- cifpanel(
title.plot = c("Fruit intake and macrovascular complications", "Zoomed-in view"),
inset.panel = TRUE,
formula = Event(t, epsilon) ~ fruitq,
data = diabetes.complications,
outcome.type = "competing-risk",
code.events = list(c(2,1,0), c(2,1,0)),
label.y = c("CIF of macrovascular complications", ""),
label.x = c("Years from registration", ""),
limits.y = list(c(0,1), c(0,0.15)),
inset.left = 0.40,
inset.bottom = 0.45,
inset.right = 1.00,
inset.top = 0.95,
inset.legend.position = "none",
legend.position = "bottom",
add.conf = FALSE
)
print(output5)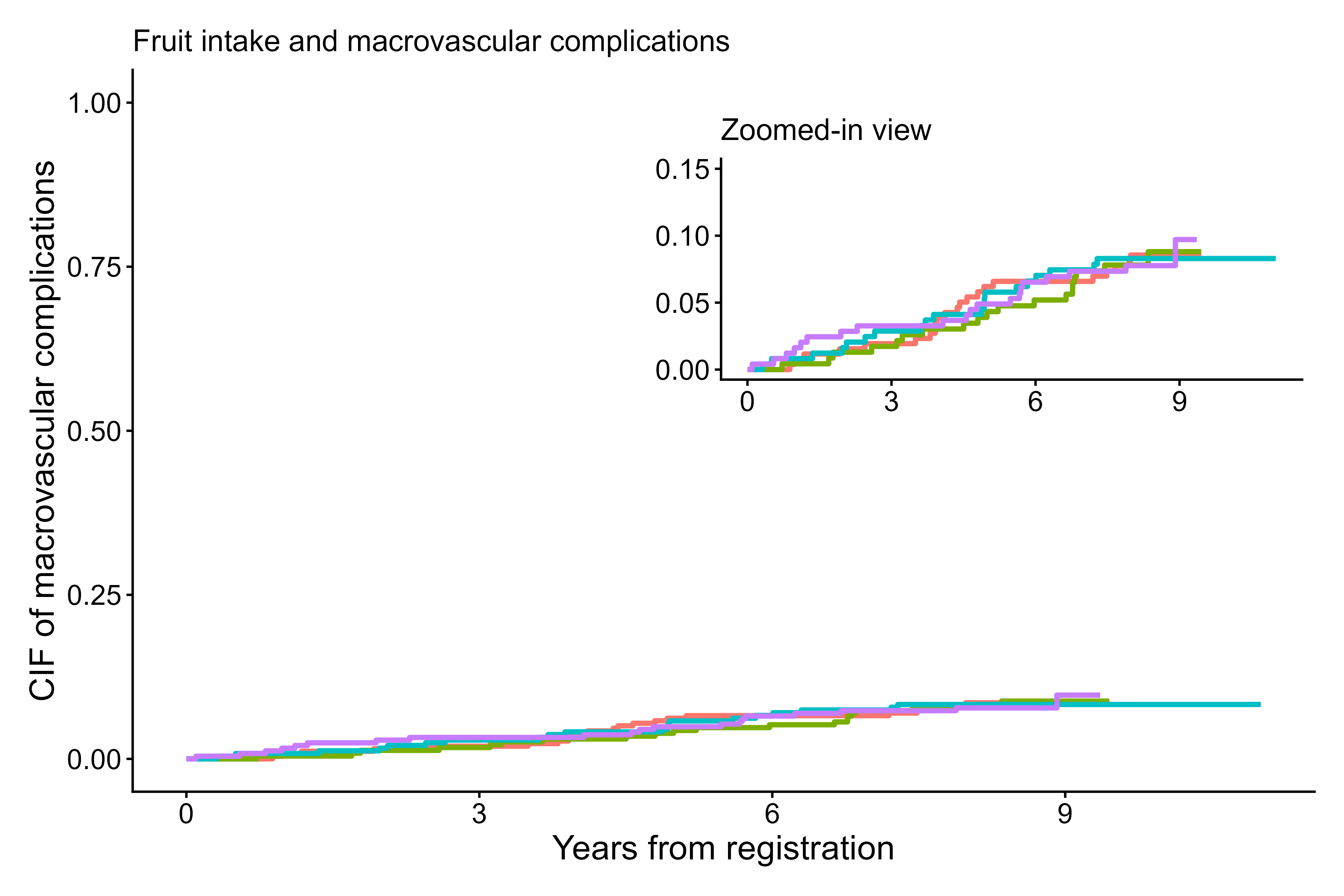
Zoomed-in-view panel using cifpanel()
Font, style and palette
cifplot(Event(t,epsilon) ~ fruitq,
data = diabetes.complications,
outcome.type = "competing-risk",
add.conf = FALSE,
font.size = 16,
font.size.risk.table = 3)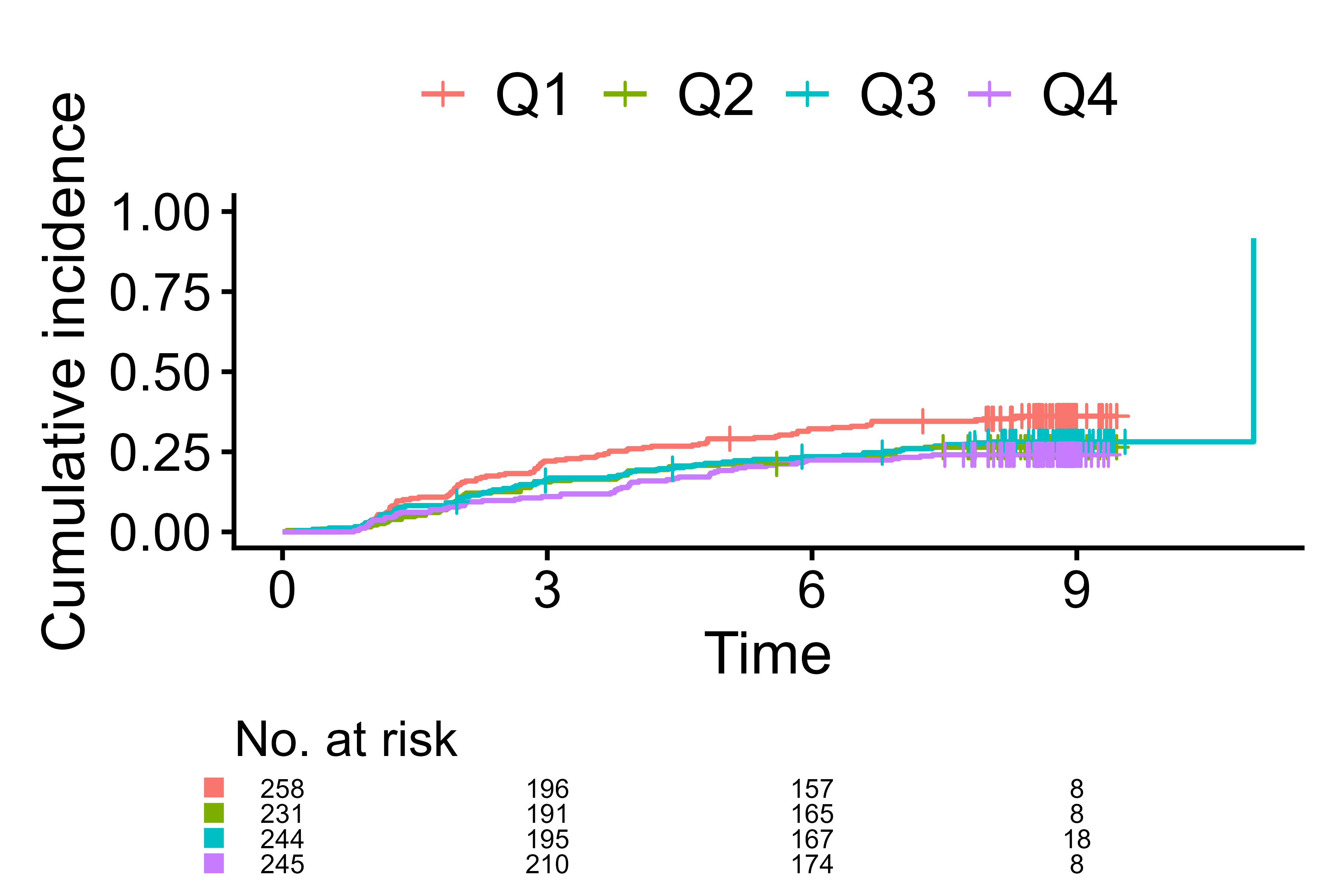
Adjust font size
cifplot(Event(t,epsilon) ~ fruitq,
data = diabetes.complications,
outcome.type = "competing-risk",
add.conf = FALSE,
style = "bold",
font.family = "serif",
palette = c("blue1", "cyan3", "navy", "deepskyblue3"))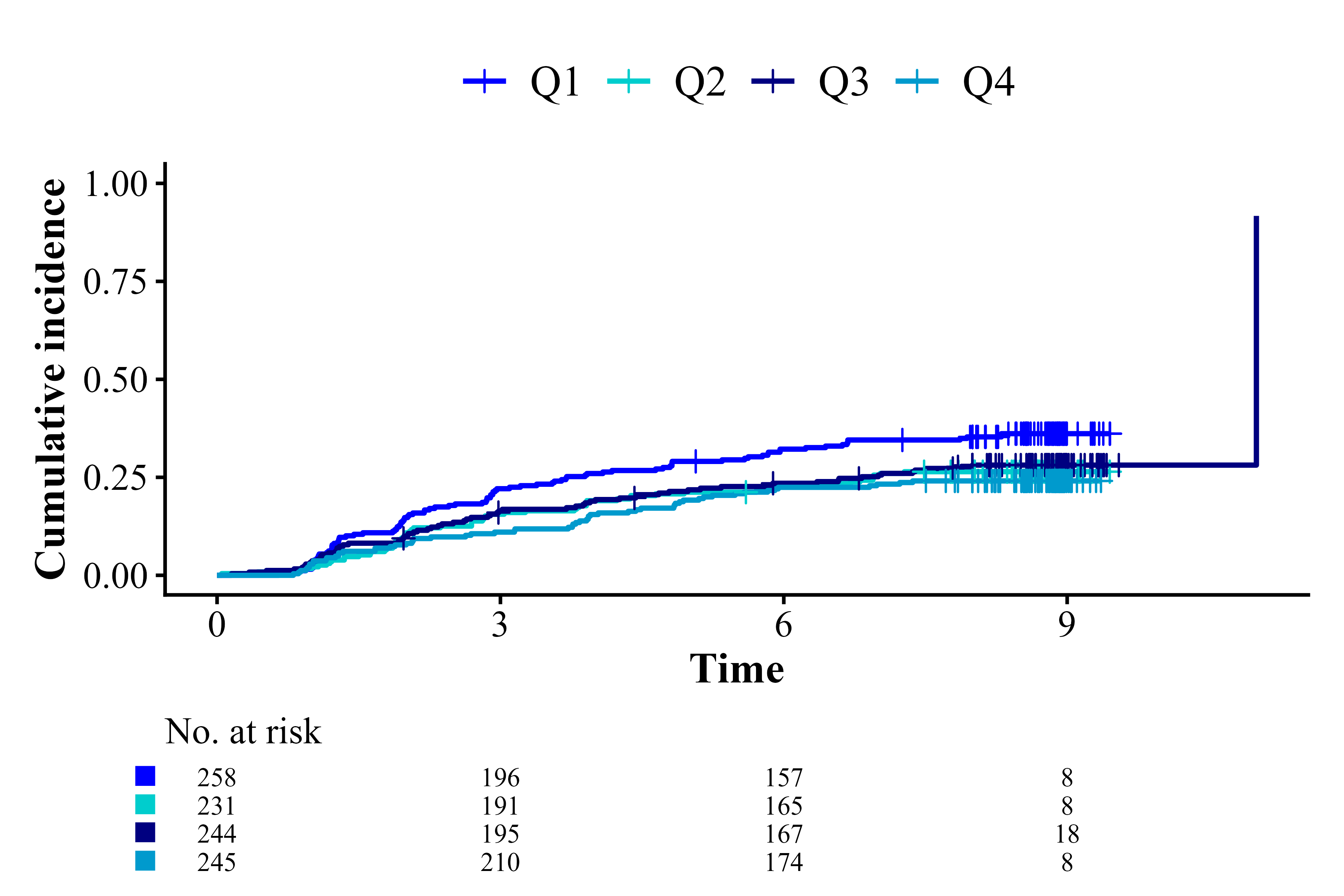
Select font family, style, linewidth and palette
cifplot(Event(t,epsilon) ~ fruitq,
data = diabetes.complications,
outcome.type = "competing-risk",
add.conf = FALSE,
style = "framed",
font.family = "serif",
linewidth = 1.3,
linetype = TRUE,
palette = c("azure4", "gray", "black", "darkgray"))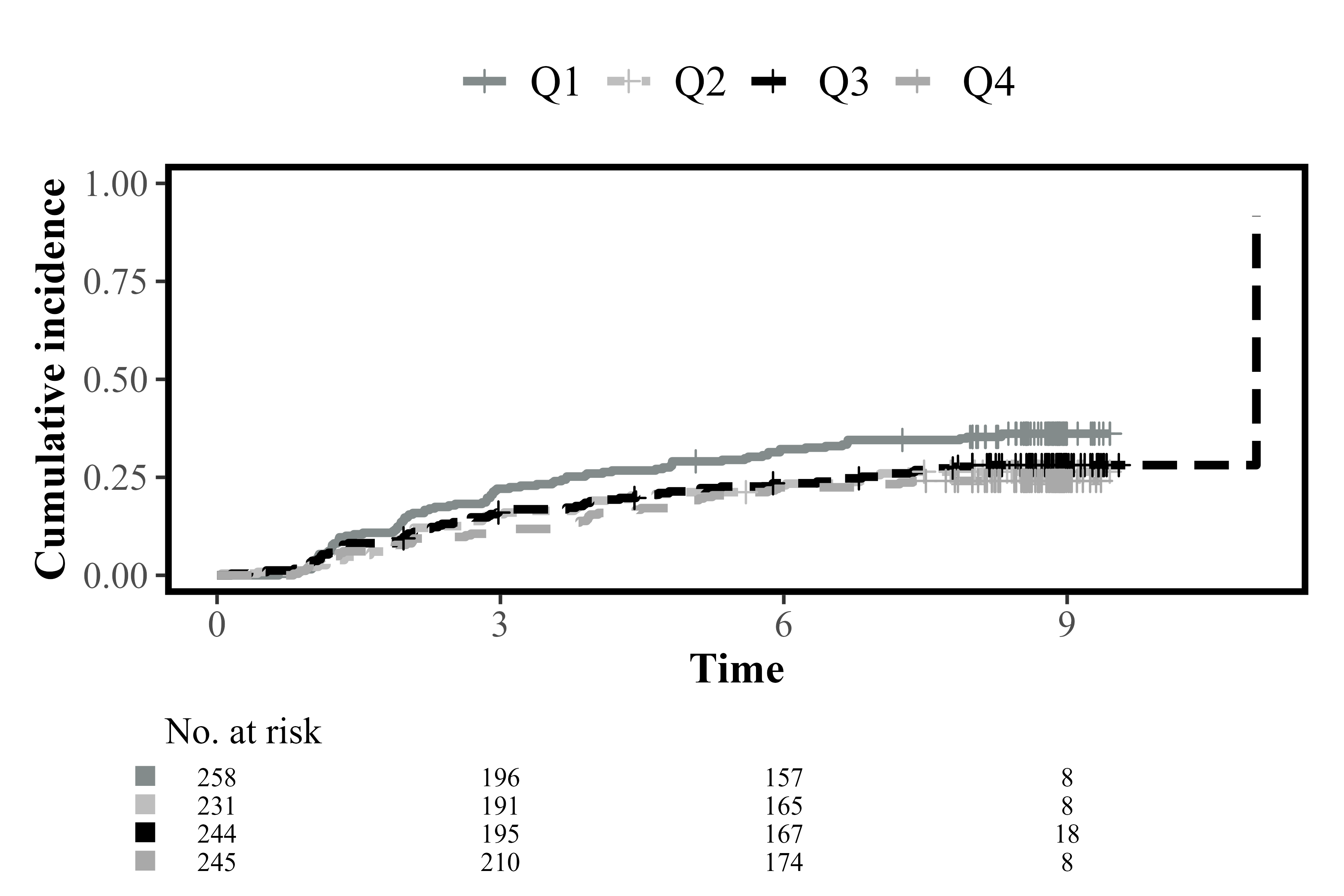
Select font family, style, linewidth and palette
cifplot(Event(t,epsilon) ~ fruitq,
data = diabetes.complications,
outcome.type = "competing-risk",
add.conf = FALSE,
style = "gray",
font.family = "mono",
palette = c("cyan3", "deepskyblue4", "darkseagreen2", "deepskyblue3"))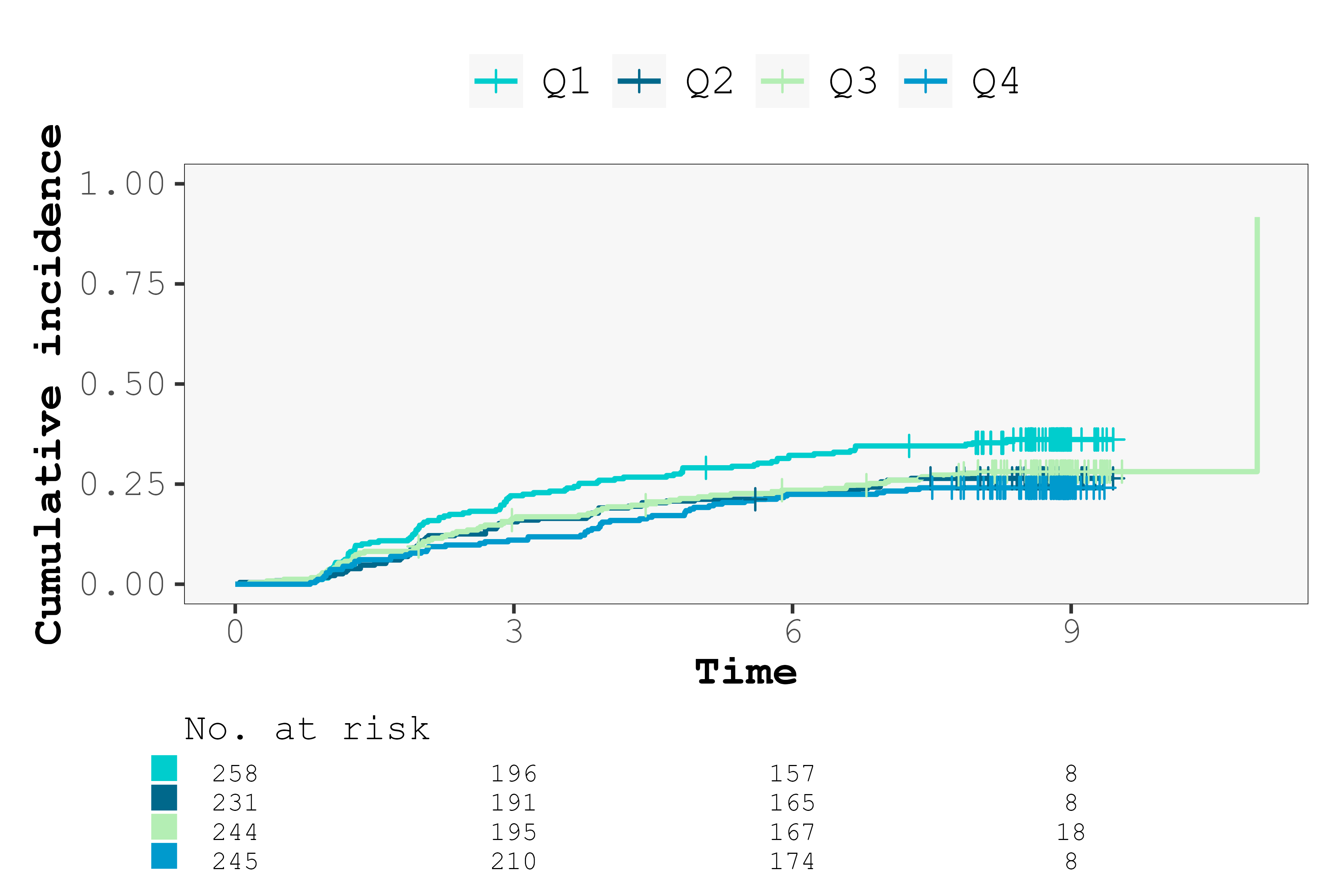
Select font family, style, linewidth and palette
cifplot(Event(t,epsilon) ~ fruitq,
data = diabetes.complications,
outcome.type = "competing-risk",
add.conf = FALSE,
style = "ggsurvfit",
font.family = "mono",
palette = c("brown", "orange", "purple", "darkgoldenrod"))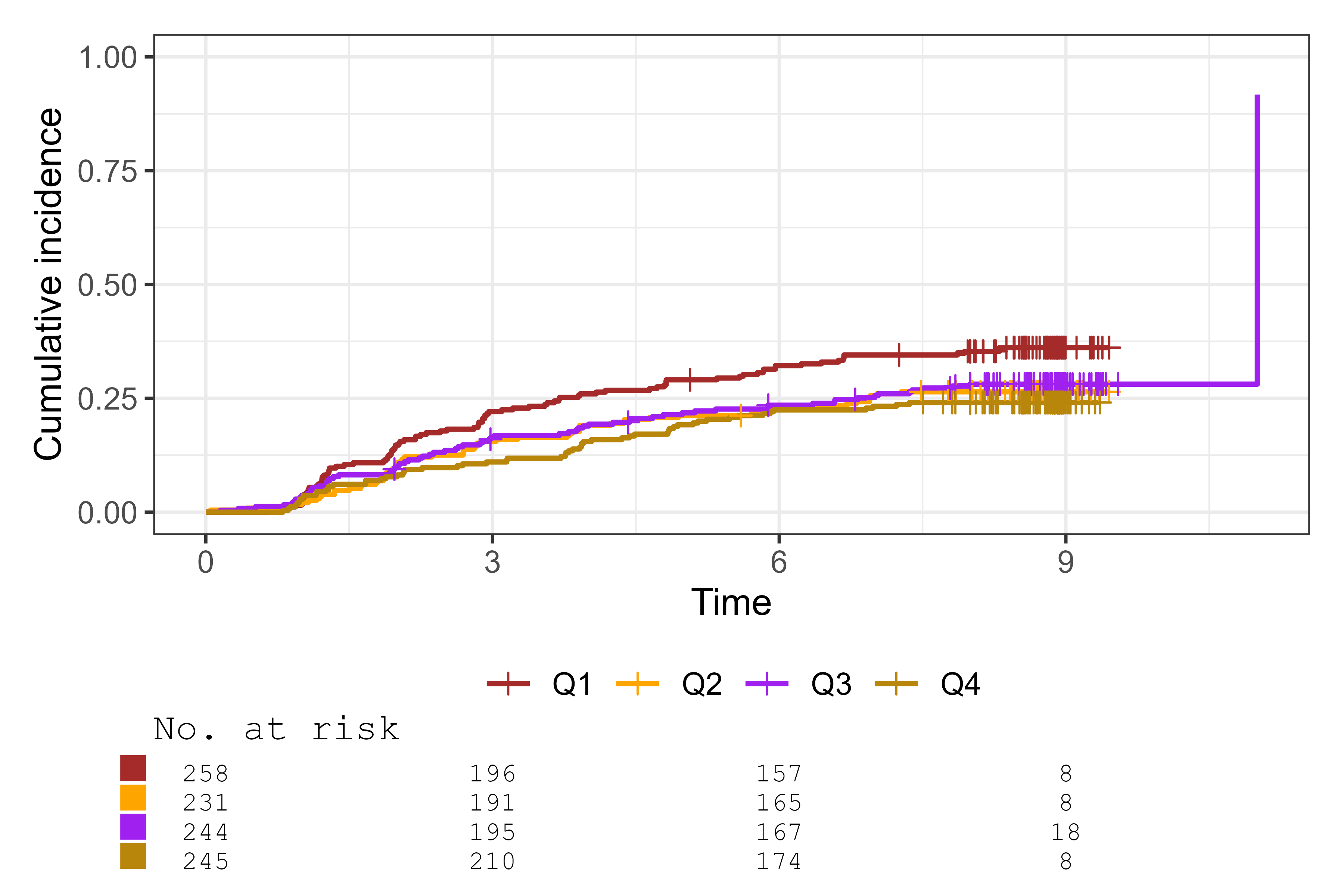
Select font family, style, linewidth and palette
Axis and label
cifplot(Event(t,d) ~ fruitq1,
data = diabetes.complications,
outcome.type = "survival",
label.y = "Survival (no complications)",
label.x = "Years from registration",
label.strata = c("High intake","Low intake"),
level.strata = c("Q2 to Q4","Q1"),
order.strata = c("Q1", "Q2 to Q4"))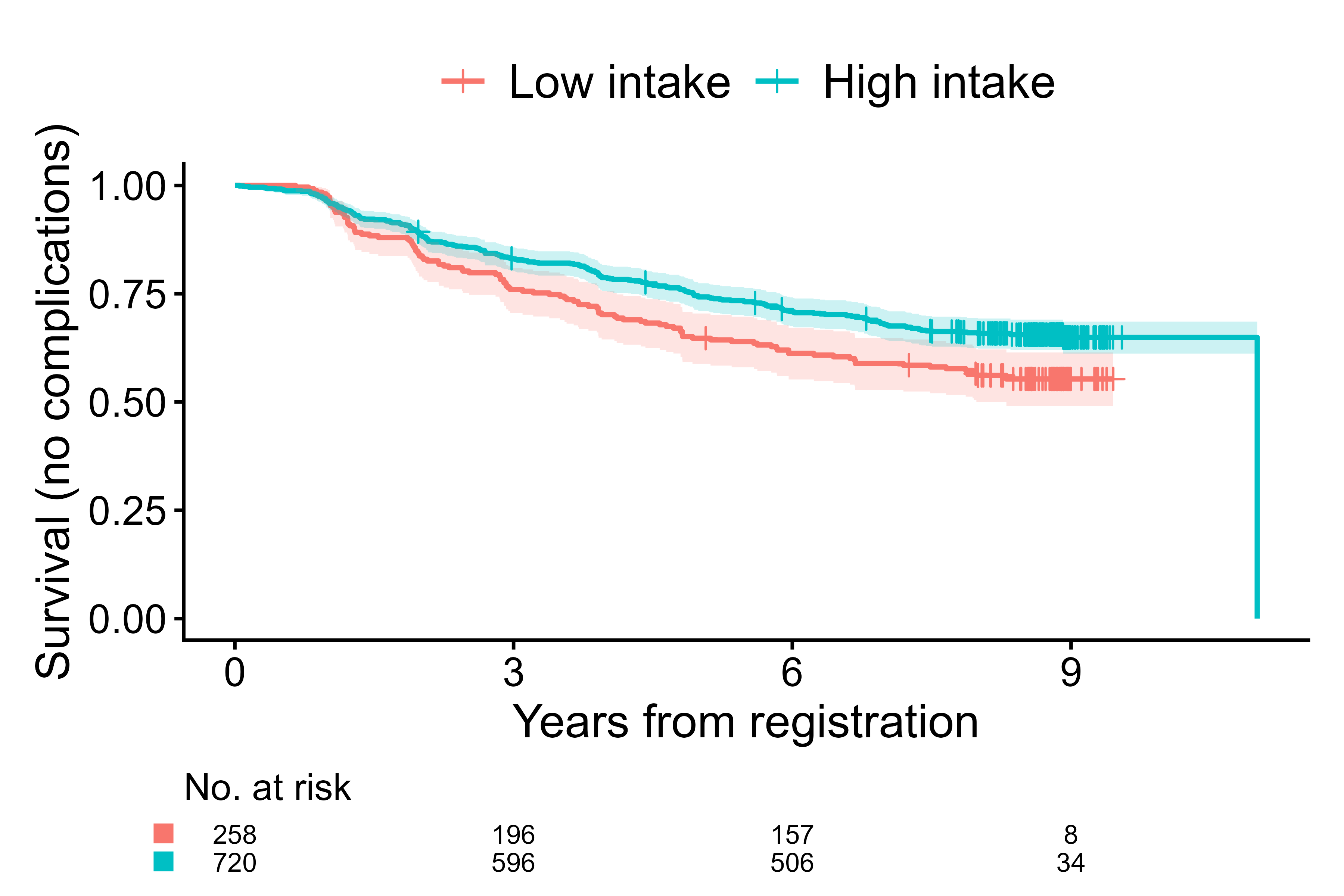
Customize labels with explicit level mapping (safe against factor order)
cifplot(Event(t,d) ~ fruitq1,
data = diabetes.complications,
outcome.type = "survival",
label.y = "Risk of diabetes complications",
label.x = "Years from registration",
label.strata = c("High intake","Low intake"),
level.strata = c("Q2 to Q4","Q1"),
order.strata = c("Q1", "Q2 to Q4"),
type.y = "risk",
limits.y = c(0, 0.5),
breaks.y = seq(0, 0.5, by=0.1),
limits.x = c(0, 8),
breaks.x = 0:8,
use.coord.cartesian = TRUE) 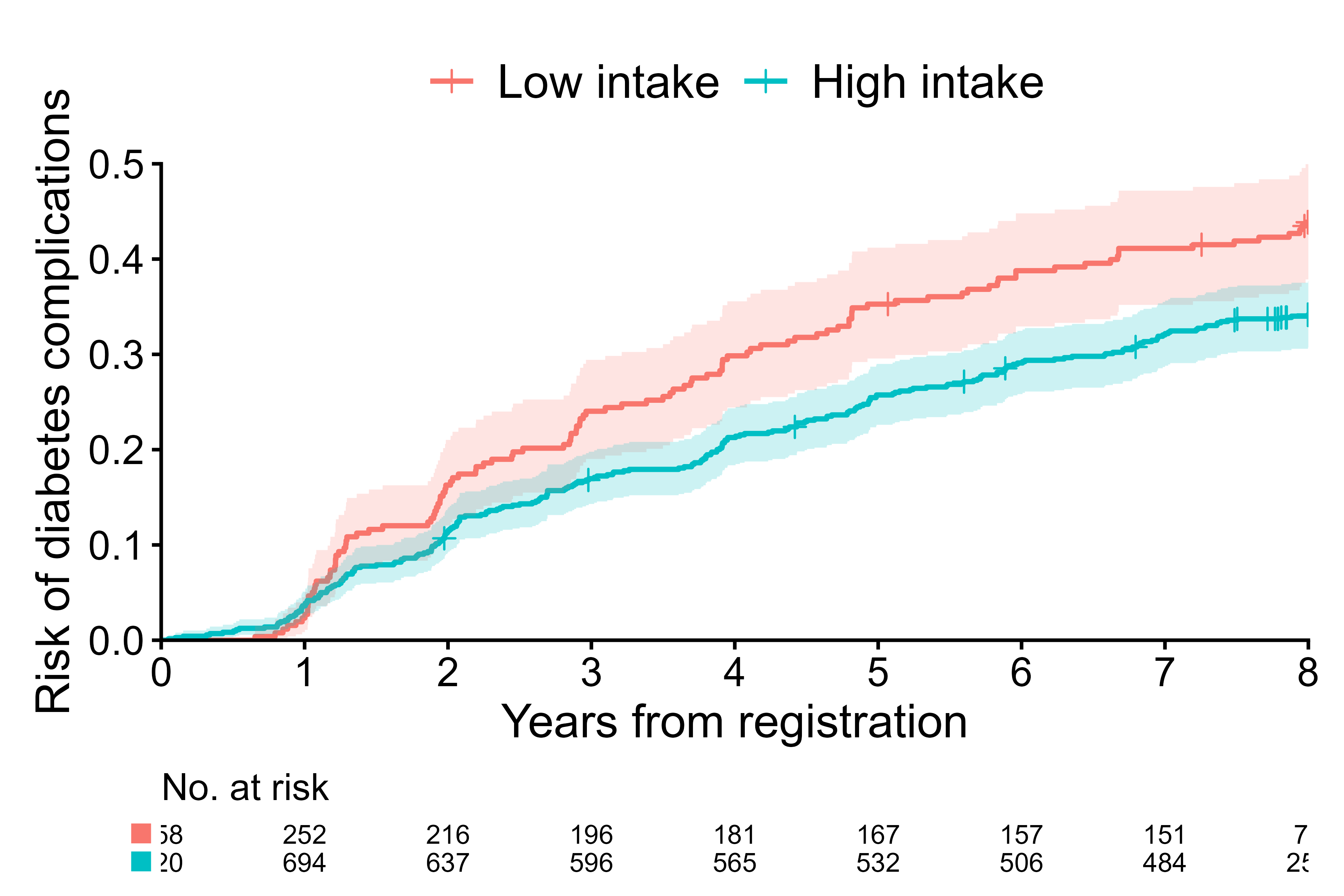
Plot 1 - survival and set axis limits